The Pool Lab is interested in how basic biological motivations are represented in the brain and how they determine the direction and content of animal behavior. In particular we focus on one of these biological drives - the mammalian pain and pain relief system. We study how distinct pain modalities are encoded in the central nervous system and how the endogenous pain relief circuits can control the perception of pain. Furthermore, we seek to take advantage of the cellular identity of these central circuit nodes to design precision gene-therapies to gain control of this system and make it therapeutically addressable.
Identification of cell-type composition and analgesia activated neuron types in brain centers implicated in pain relief.
Mapping and dissection of mammalian pain relief circuits
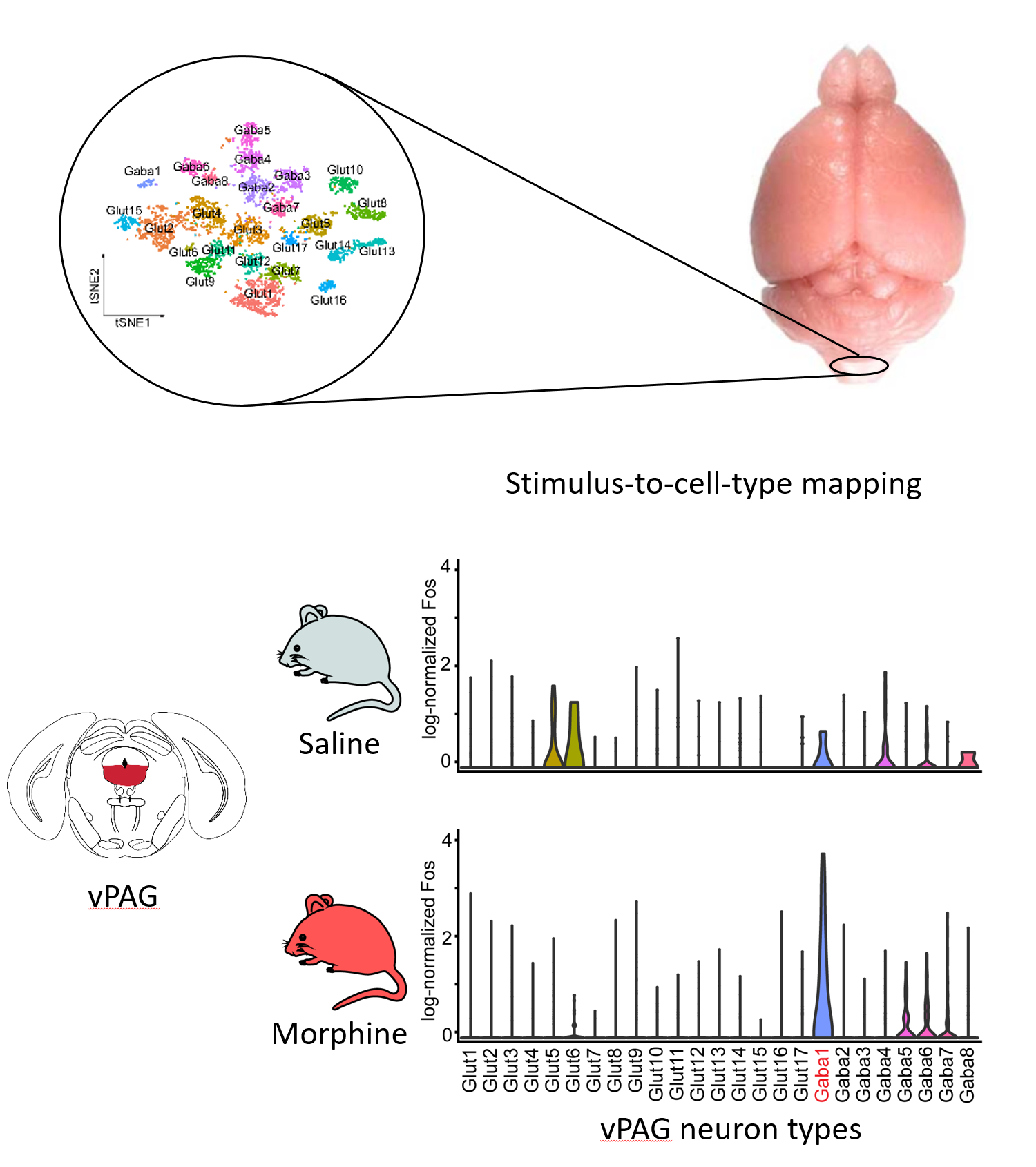
Previous work has identified a dozen brain nuclei in the brainstem, thalamus and cortex that can impart profound pain relief upon crude electric stimulation or localized opioid delivery demonstrating the presence of cell populations capable of conveying analgesia. Although the general anatomy of this system has been known for decades, the precise cell types responsible for these effects and means to selectively target them are mostly unknown. By using a single-cell profiling based stimulus-to-cell-type mapping approach that we recently demonstrated, we have uncovered a number of analgesic state activated and sensitive neuron types in the brain. We aim to identify their full repertoire and characterize which elements of this circuit can gate the expression of specific pain states.
Mapping the central representation of pain modalities
Recent work has elegantly elucidated the molecular identity and functional contribution of different pain sensory neurons in the sensory ganglia. The cellular identity and functional role of central pain circuit nodes however is much less clear. In particular, we have a limited understanding how the sensation of pain is translated into adaptive avoidance and recuperative behaviors, associative learning and how it is transformed into an unpleasant subjective experience. Furthermore, to which extent different acute pain modalities and chronic pain states engage shared or distinct supra-spinal pain pathways is incompletely understood. We are taking an unbiased approach for identifying central pain activated neuron types through spatial transcriptomic and scRNA-seq based stimulus-to-cell-type mapping focusing on brainstem, thalamic and cortical regions harboring pain activated neurons. We seek to elucidate the functional contribution of identified pain activated neurons to shaping adaptive pain behaviors in acute and chronic pain states through viral cell-type-specific manipulations and in vivo calcium imaging. These experiments will reveal which pain modalities engage which central pain networks and pinpoint candidate circuit nodes that could serve as targets for controlling the experience of specific types of pain.
High-throughput mapping of distinct pain-state engaged neuron types in central pain relays with single molecule spatial transcriptomics (MERFISH).
Cell-type-targeted gene therapies
We seek to leverage the uncovered local cell-type-defining genetic programs of the pain and analgesia circuit nodes to design precision gene therapies to make these circuits selectively therapeutically addressable. To this end, in preliminary work, we have developed a high efficiency gene editing based gene therapy solution for functional reprogramming of specific cell types in vivo. We will deploy this and competing strategies for precision control of specific pain and analgesia circuit nodes to achieve targeted and side-effect free induction of pain relief in a diverse array of pain conditions. Importantly, this provides a path to controlling this system outside specialized model organisms and paves the way for human applications. Thereby we seek to transform how heterogeneous pain conditions that currently remain poorly addressed can be tackled.